|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Individual contracting cardiomyocytes are large cells between 20 µm (atrial cardiomyocytes) and 140 µm
 Figure 18-9
Organization of cardiomyocytes. Fifty percent of cardiomyocyte
volume is made up of myofibrils; the remainder consists of mitochondria, nucleus,
sarcoplasmic reticulum, and cytosol.
Figure 18-9
Organization of cardiomyocytes. Fifty percent of cardiomyocyte
volume is made up of myofibrils; the remainder consists of mitochondria, nucleus,
sarcoplasmic reticulum, and cytosol.
The sarcolemma, or the outer plasma membrane, separates the intracellular and extracellular space. It surrounds the cardiomyocyte and invaginates into the myofibrils through an extensive tubular network known as transverse or T tubules, and it also forms specialized intercellular junctions between cells.[18] [19]
Transverse or T tubules are in close proximity to an intramembrane system and the SR, which plays an important role in calcium metabolism that is critical in excitation-contraction coupling of the cardiomyocyte. The SR can be further divided into the longitudinal (or network) and the junctional SR. The longitudinal SR is involved in the uptake of calcium for initiation of relaxation. The junctional SR contains the large calcium release channels (ryanodine receptors) that release SR calcium stores in response to depolarization-stimulated calcium influx through the sarcolemmal calcium channels. The ryanodine receptors not only are calcium release channels but also form the scaffolding proteins that anchor many of the key regulatory proteins.[20]
Mitochondria are found immediately beneath the sarcolemma, wedged between myofibrils within the cell. They contain enzymes that promote the generation of adenosine triphosphate (ATP), and they are the energy powerhouse for the cardiomyocyte. In addition, mitochondria can also function to accumulate calcium and thereby contribute to regulation of the cytosolic calcium concentration. Nearly all of the genetic information is found within the centrally located nucleus. The cytosol is the fluid-filled microenvironment within the sarcolemma, exclusive of the organelles and the contractile apparatus and proteins.
Cardiac muscle cells contain three different types of intercellular junctions: gap junctions, "spot" desmosomes, and
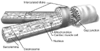 Figure 18-10
Sarcolemma that envelops cardiomyocytes becomes highly
specialized to form the intercalated disks where ends of neighboring cells are in
contact. The intercalated disks consist of gap junctions and "spot" and "sheet"
desmosomes.
Figure 18-10
Sarcolemma that envelops cardiomyocytes becomes highly
specialized to form the intercalated disks where ends of neighboring cells are in
contact. The intercalated disks consist of gap junctions and "spot" and "sheet"
desmosomes.
The conducting cardiomyocytes, or Purkinje cells, are specialized cells for conducting propagated action potentials. These cells have a low content of myofibrils and a prominent nucleus and contain an abundance of gap junctions. Cardiomyocytes can be functionally separated into (1) the excitation system, (2) excitation-contraction-coupling, and (3) the contractile system.
The cellular action potential originating in the specialized conduction tissue is propagated to individual cells where it initiates the intracellular event that leads to contraction of the cell through the sarcolemmal excitation system.
Ion fluxes across plasma membranes result in depolarization (attaining a less negative membrane potential) and repolarization (attaining a more negative membrane potential). They are mediated by membrane proteins with ion-selective pores. Because these ion channel proteins open and close the pores in response to changes in membrane potential, the channels are voltage gated. In the heart, sodium, potassium, calcium, and chloride channels have been found to contribute to the action potential.
The types of action potential in the heart can be separated into two categories: fast-response action potentials, which are found in the His-Purkinje system and atrial or ventricular cardiomyocytes, and slow-response action potentials, which are found in the pacemaker cells in the SA and AV nodes. A typical tracing for an action potential
In contrast, during diastole (phase 4), pacemaker cells that show slow-response action potentials have the capability of spontaneous diastolic depolarization and generate the automatic cardiac rhythm. Pacemaker currents during phase 4 are the result of an increase in the three inward currents and a decrease in the two outward currents. The three inward currents that contribute to spontaneous
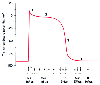 Figure 18-11
Phases of cellular action potentials and major associated
currents in ventricular myocytes. The initial phase zero spike and overshoot (1)
are caused by a rapid inward Na current, the plateau phase (2) by a slow Ca current
through L-type Ca channels, and repolarization (phase 3) by outward K currents.
Phase 4, the resting potential (Na efflux, K influx), is maintained by Na-K-ATPase.
The Na-Ca exchanger is mainly responsible for Ca extrusion. In specialized conduction
system tissue, spontaneous depolarization takes place during phase 4 until the voltage
resulting in opening of the Na channel is reached. (From LeWinter MM, Osol
G: Normal physiology of the cardiovascular system. In
Fuster V [ed]: Hurst's The Heart, 10th ed. New York, McGraw-Hill, 2001, pp 63–94.)
Figure 18-11
Phases of cellular action potentials and major associated
currents in ventricular myocytes. The initial phase zero spike and overshoot (1)
are caused by a rapid inward Na current, the plateau phase (2) by a slow Ca current
through L-type Ca channels, and repolarization (phase 3) by outward K currents.
Phase 4, the resting potential (Na efflux, K influx), is maintained by Na-K-ATPase.
The Na-Ca exchanger is mainly responsible for Ca extrusion. In specialized conduction
system tissue, spontaneous depolarization takes place during phase 4 until the voltage
resulting in opening of the Na channel is reached. (From LeWinter MM, Osol
G: Normal physiology of the cardiovascular system. In
Fuster V [ed]: Hurst's The Heart, 10th ed. New York, McGraw-Hill, 2001, pp 63–94.)
During the cardiac action potential, movement of Ca2+ into the cell and Na+ out of the cell creates an ionic imbalance. The Na+ -Ca2+ exchanger restores cellular ionic balance by actively transporting Ca2+ out of the cell against a concentration gradient while moving Na+ into the cell in an energy-dependent manner.
The structures that participate in cardiac excitation-contraction coupling (ECC) include the sarcolemma, transverse tubules, SR, and the myofilaments ( Fig. 18-12A ).[23] The process of ECC begins with depolarization of the plasma membrane and the spread of electrical excitation along the sarcolemma of cardiomyocytes. Calcium enters through plasma membrane channels concentrated at the T tubules, and such entry triggers the release of calcium from the SR, which in turn stimulates myofibrillar contraction. [24]
The ubiquitous second messenger calcium is the key player in cardiac ECC ( Fig. 18-12B ).[23] The cycling of calcium within the structures that participate in ECC initiates and terminates contraction. Activation of the contractile system depends on the increase in free cytosolic calcium and its subsequent binding to contractile proteins. The influx of calcium through the voltage-gated L-type calcium channels (dihydropyridine receptors) causes an initial small increase in intracellular calcium. The local increase in Ca2+ concentration as a result of the entry of Ca2+ through the voltage-gated L-type calcium channels activates the calcium release channels (ryanodine receptors), induces further release of calcium from the subsarcolemmal cisternae in the SR, and thus leads to a large increase in intracellular calcium. The increase in intracellular calcium, however, is transient inasmuch as calcium is removed by (1) active uptake by the SR calcium pump adenosine triphosphatase (ATPase), (2) extrusion of calcium from the cytosol by the Na+ -Ca2+ exchanger, and (3) binding of calcium to proteins.[25]
The SR provides the anatomic framework and is the major organelle for the cycling of calcium. It is the depot for intracellular calcium stores. The cyclic release plus reuptake of calcium by the SR regulates the cytosolic calcium concentration and couples excitation to contraction. The physical proximity between the L-type calcium channels and the ryanodine receptors at the SR membrane makes calcium-induced calcium release occur easily. The "foot" region of the ryanodine receptor is the part that extends from the SR membrane to the T tubules, where the L-type calcium channels are located.[16] [25] [26]
The SR is also concerned with the reuptake of calcium that initiates relaxation or terminates contraction. Sarcoplasmic reticulum ATPase (SERCA) is the ATP-dependent pump that actively pumps the majority of the calcium back into the SR after its release. SERCA makes up
 Figure 18-12
A, Diagram depicting the
components of cardiac excitation-contraction coupling. Calcium pools are noted in
bold letters. B, Extracellular (arrows
A, B1, B2) and intracellular calcium flux (arrows,
C, D, E, F, G) is shown. The thickness of the arrows
indicates the magnitude of the calcium flux, the vertical orientations describe their
energetics: downward-pointing arrows represent passive
calcium flux, and upward pointing arrows represent
energy-dependent calcium transport. Calcium entering the cell from extracellular
fluid through L-type calcium channels triggers calcium release from the sarcoplasmic
reticulum (SR). Only a small portion directly activates the contractile proteins
(A1). Arrow B1 depicts active transport of calcium
into the extracellular fluid by means of the plasma membrane pump ATPase and the
Na+
-Ca2+
exchanger. Sodium that enters the cell in exchange
for calcium (dashed line) is pumped out of the cytosol
by the sodium pump. SR regulates calcium efflux from the subsarcolemmal cisternae
(arrow C) and calcium uptake into the sarcotubular
network (arrow D). Arrow G
represents calcium that diffuses within the SR. Calcium binding to (arrow
E) and dissociation from (arrow F) high-affinity
calcium binding sites of troponin C activate and inhibit interactions of the contractile
proteins. Arrow H depicts calcium movement into
and out of mitochondria to buffer the cytosolic calcium concentration. (From
Katz AM: Calcium fluxes. In Physiology of the Heart,
3rd ed. Philadelphia, Lippincott-Raven, 2001, pp 232–233.)
Figure 18-12
A, Diagram depicting the
components of cardiac excitation-contraction coupling. Calcium pools are noted in
bold letters. B, Extracellular (arrows
A, B1, B2) and intracellular calcium flux (arrows,
C, D, E, F, G) is shown. The thickness of the arrows
indicates the magnitude of the calcium flux, the vertical orientations describe their
energetics: downward-pointing arrows represent passive
calcium flux, and upward pointing arrows represent
energy-dependent calcium transport. Calcium entering the cell from extracellular
fluid through L-type calcium channels triggers calcium release from the sarcoplasmic
reticulum (SR). Only a small portion directly activates the contractile proteins
(A1). Arrow B1 depicts active transport of calcium
into the extracellular fluid by means of the plasma membrane pump ATPase and the
Na+
-Ca2+
exchanger. Sodium that enters the cell in exchange
for calcium (dashed line) is pumped out of the cytosol
by the sodium pump. SR regulates calcium efflux from the subsarcolemmal cisternae
(arrow C) and calcium uptake into the sarcotubular
network (arrow D). Arrow G
represents calcium that diffuses within the SR. Calcium binding to (arrow
E) and dissociation from (arrow F) high-affinity
calcium binding sites of troponin C activate and inhibit interactions of the contractile
proteins. Arrow H depicts calcium movement into
and out of mitochondria to buffer the cytosolic calcium concentration. (From
Katz AM: Calcium fluxes. In Physiology of the Heart,
3rd ed. Philadelphia, Lippincott-Raven, 2001, pp 232–233.)
Once taken up into the SR, calcium is stored until it is released during the next cycle. Calsequestrin and calreticulin are two storage proteins in the SR. Calsequestrin is a highly charged protein located in the cisternal component of the SR near the T tubules. Because it lies close to the calcium release channels, the stored calcium can be quickly discharged for release once the calcium release channels are stimulated.
Cytosolic calcium can also be removed by extrusion through the sarcolemmal calcium pump and the activity of the sodium-calcium exchanger. An important "sensor" and regulator of intracellular calcium is the protein calmodulin.[18]
The basic working unit of contraction is the sarcomere. A sarcomere is defined as the distance between Z lines (Z is an abbreviation for the German Zuckung, or "contraction"), which join the sarcomeres in series. Each sarcomere consists of a central A band that is separated by half of an I band from the Z lines on each side because the
Familial hypertrophic cardiomyopathy is an inherited autosomal dominant sarcomeric disease[29] that is the most common cause of sudden death in otherwise healthy individuals. Its clinical features are left ventricular hypertrophy and myocyte/myofibrillar disarray. Mutations in at least eight different genes encoding sarcomere proteins have been identified to be the molecular basis for the disorder. These genes are β-cardiac myosin heavy chain, cardiac troponin T, α-tropomyosin, cardiac myosin binding protein C, essential or regulatory myosin light chain, cardiac troponin I, α-cardiac actin, and titin.[29]
The contractile apparatus within the cardiomyocyte consists of contractile and regulatory proteins.[18] [30] [31] The thin filament actin and the thick filament myosin are the two principal contractile proteins. Actin contains two helical chains. Tropomyosin, a double-stranded α-helical regulatory protein, winds around the actin array and forms the backbone for the actin thin filament. The myosin thick filament is made up of 300 myosin molecules. Each myosin molecule has two functional domains: the body or filament and the bilobar myosin head. The myosin head is composed of one heavy chain and two light chains. The heavy head chain has two domains: the larger one interacts with actin at the actin cleft and has an ATP-binding pocket where myosin
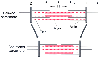 Figure 18-13
The basic unit for contraction is the sarcomere. A contracted
and a relaxed sarcomere is depicted. Z lines are located at the ends of the sarcomere.
The A band is the site of overlap between myosin and actin filaments. The I band
is located on either side of the A band and contains only actin filament. The H
zone is located in the center of the A band, and only myosin is present.
Figure 18-13
The basic unit for contraction is the sarcomere. A contracted
and a relaxed sarcomere is depicted. Z lines are located at the ends of the sarcomere.
The A band is the site of overlap between myosin and actin filaments. The I band
is located on either side of the A band and contains only actin filament. The H
zone is located in the center of the A band, and only myosin is present.
At rest, cross-bridge cycling and generation of force do not occur because either the myosin heads are blocked from physically reacting with the thin filament or they are only weakly bound to actin ( Fig. 18-14 ).[16] Cross-bridge cycling is initiated on binding of calcium to TnC, which increases the TnC-TnI interaction and decreases the inhibitory TnI-actin interaction. These events that ensue from the binding of Ca2+ to TnC lead to conformational changes in tropomyosin and permit attachment of the myosin head to actin. Cross-bridging involves detachment of the myosin head from actin and reattachment of myosin to another actin on hydrolysis of ATP by myosin ATPase. ATP binding to the nucleotide pocket of the myosin head leads to the activation of myosin ATPase,[26] [27] [28] ATP hydrolysis, and changes in the configuration of the myosin head, thus facilitating the binding of myosin head to actin and generation of the power stroke of the myosin head. Based on this model, it is apparent that the rate of cross-bridge cycling is dependent on the activity of myosin ATPase.[31] The turnoff of cross-bridge cycling is largely initiated by the fall in cytosolic calcium.
Myocyte relaxation is an energy-dependent process because the restoration of cytosolic calcium to resting levels requires the expenditure of ATP. The fall in cytosolic calcium occurs through the active reuptake of calcium into the SR by SERCA and extrusion of calcium by the Na+ -Ca2+ exchanger. This activity results in the release of Ca2+ binding from TnC and separation of the myosinactin cross-bridge. Myocyte relaxation is dependent on the kinetics of cross-bridge cycling, the affinity of Ca2+ for TnC, and the activity of the calcium reuptake mechanisms. Relaxation is enhanced by increased kinetics of cross-bridge cycling, decreased Ca2+ affinity for TnC, and increased activity of calcium reuptake mechanisms.[25]
Titin is a giant stringlike protein that acts as the third filament within the sarcomere. A single titin molecule spans half the sarcomere. Structurally, titin consists of an inextensible anchoring segment and an extensible elastic segment. Its two main functions involve muscle assembly and elasticity. Titin is the principal determinant of the passive properties of the myocardium at low ventricular volume. [32]
The Frank-Starling relationship states that an increase in end-diastolic volume results in enhanced systolic function.[33] [34] At the cellular level, the key component for the Frank-Starling relationship is a length-dependent shift in Ca2+ sensitivity. [35] [36] [37] Several possible mechanisms for this change in Ca2+ sensitivity have been implicated,
 Figure 18-14
Molecules of the contractile system. (From Opie
LH: Myocardial contraction and relaxation. In The
Heart. Physiology from Cell to Circulation, 3rd ed. Philadelphia, Lippincott-Raven,
1998, pp 209–231.)
Figure 18-14
Molecules of the contractile system. (From Opie
LH: Myocardial contraction and relaxation. In The
Heart. Physiology from Cell to Circulation, 3rd ed. Philadelphia, Lippincott-Raven,
1998, pp 209–231.)
The cytoskeleton is the protein framework within the cytoplasm that links, anchors, or tethers structural components inside the cell.[17] [18] Microfilament (actin filament), microtubules, and intermediate filaments are three classes of cytoskeleton proteins found in the cytoplasm. Microfilament proteins are actin filaments, either sarcomeric or cortical, depending on their location. Sarcomeric actin filaments are the thin filaments in the contractile machinery that have already been described. Cortical actin filaments are found below the plasma membranes at the cell surface and are linked to several other microfilament proteins, including dystrophin, vinculin, and ankyrin. Microtubules assemble by polymerization of the α- and β-dimers of tubulin. They play a major role in intracellular transport and cell division.[38] Attachment of the ends of microtubules to cellular structures causes the microtubules to expand and contract, thereby pulling and pushing these structures around the cell. The intermediate filaments are relatively insoluble. They have been demonstrated to be important in normal mitochondrial function and behavior. The desmin intermediate filament in cardiomyocytes connects the nucleus to the plasma membrane and is important in transmission of the stress and strain of contractile force between cells.[39] The cytoskeleton provides the organization of microenvironments within the cell for enzyme/protein activity and interaction.
Whereas familial hypertrophic cardiomyopathy is a genetic sarcomeric disease, familial dilated cardiomyopathy
|
|
|
|
|
|
|
|
|
|
|
|
|