RESEARCH AND DETAILED INFORMATION
Detailed examination of the scientific literature that shapes
and informs the practice of anesthesia is best carried out by using the MEDLINE database.
As noted earlier, MEDLINE is a bibliographic database that contains citations to
more than 14 million biomedical articles. Until recently, MEDLINE could be accessed
only by library professionals, who used a rather primitive search program and a very
precise grammar and vocabulary. The NLM now provides free, global access to MEDLINE
through a relatively easy-to-use, web-centric interface at PubMed.gov. The current
user interface is illustrated in Figure
80-3
.
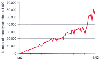 Figure 80-4
Growth in the use of PubMed. As of November 2003, there
were approximately 2 million PubMed searches per day. (Data courtesy of
PubMed.gov.)
Figure 80-4
Growth in the use of PubMed. As of November 2003, there
were approximately 2 million PubMed searches per day. (Data courtesy of
PubMed.gov.)
About 2 million PubMed searches take place per day ( Fig.
80-4
). Entrez is the name of the software system used to translate a human
user's search terms into the specific terms and grammar most likely to produce the
desired results from MEDLINE. Creating a simple interface to a massive database
is no easy task for a number of reasons, most prominent being the lack of a common
standard vocabulary among the global authors of the literature. The NLM expended
considerable effort to create a common dictionary for medical words and phrases ("MeSH"),
and Entrez tries to expand the users' search terms so that synonyms, local terms,
obsolete terms, and abbreviations are translated into those used internally in the
MEDLINE database. Entrez also combines the medical literature together with DNA
and protein sequence databases, three-dimensional protein structure and protein domain
data, population study data sets, expression data, assemblies of complete genomes,
and taxonomic information into a tightly interlinked, cross-referenced system.
Medical Search Strategies
Finding relevant publications among the many millions of entries
in MEDLINE involves matching specific words (keywords) or medical topic names from
the user to specific entries that contain these words. Generally, keywords are chosen
to reflect the key concepts being sought or the names of specific authors, or both.
Searching a computer database usually involves making a trade-off between the sensitivity
and specificity of the results. A search designed to be highly specific runs the
risk of missing some relevant citations in reporting its results, whereas a sensitive
search may produce an overwhelming number of citations, many of which will be irrelevant
to the immediate question posed. In the interest of efficiency, a MEDLINE user should
start with a relatively specific question, initiate the search with the chosen keywords,
and be prepared to loosen some of the constraining keywords if the results of the
initial search are inadequate. For example, a search for drug toxicity reports should
start with the drug name, the sought type of toxicity, and the target population
of interest. An illustrative case might be a search for the nature of ocular complications
as a result of the use of succinylcholine in patients with open-globe trauma. A
search using just the keyword succinylcholine produces 6444 citations, clearly too
sensitive and not at all specific. Search with "succinylcholine ocular" produces
65 results, a manageable number, but most of the results can be seen to refer to
closed-globe conditions. Search with a more specific set of keywords, such as "succinylcholine
open globe," and 13 highly relevant citations are produced. Entrez allows such combinations
of keywords under the assumption that a simple unpunctuated list of words requires
that each word be present somewhere in a citation, though not necessarily adjacent
to the other words in the list. This particular relationship of search words is
also known as the AND Boolean operator. Also available in Entrez is the OR operator,
which will match a citation if a word on either side of the OR is matched, and the
NOT operator, which will reject citations that include the word following the NOT.
A phrase in which words must be adjacent or in a specific order can be specified
by enclosing the phrase in double quotes, for example, "endotracheal intubation."
Note that Boolean operators must be fully capitalized when used in an Entrez search;
otherwise they will be treated as simple keywords. Entrez can also simultaneously
find up to 150 words that start with the same root by using the truncation operator
"*"; for example, anesth* will match anesthesia, anesthetic, anesthesiologist,
and so forth.
Returning briefly to expand on the conceptual structure of a database,
consider that MEDLINE is a collection of entities, specifically, bibliographic citations.
As in any database, the actual information in each citation is divided into a set
of predefined types or fields. The fields in a database of hospital patients could
consist of patient name, medical record number, date of birth, admitting physician,
and other fields. Like any database, each bibliographic citation in MEDLINE is composed
of fields (over 30 actually), including authors, title, abstract, publication type,
authors' address, publication date, journal title, issue number, page numbers, language,
MeSH topics and words, and others. One of the powerful search features of the PubMed
database is its use of field tags. Entrez allows a search to be specified by the
contents of specific fields, which greatly increases the precision of a search.
For example, a search for English language review articles in American anesthesia
journals on pain could be written:
Pain[majr] eng[LA] Review[PT] Anesth*[TA]
Field tags are usually two letter abbreviations enclosed in square
brackets. In this example, for a citation to match, pain is required to be a major
MeSH topic, the language must be English, the publication type must be a review,
and the word root Anesth* must appear in the journal name. Because there are
quite a number of field tag abbreviations, the PubMed web page provides a shortcut
hyperlink called "Limits" that provides a search page with the most common field
tags made available in simple drop-down menus.
A complete review on using MEDLINE and its services can be found
at www.nlm.nih.gov/bsd/PubMed_tutorial/m1001.html.
The NLM also maintains its own digital archive of the electronic
(nonprint) life sciences journal literature in PubMed Central. Here, cooperating
publishers voluntarily
contribute their content, which is then indexed and maintained by the NLM. Another
interesting database maintained for public access by the NLM is the PubMed Bookshelf.
This freely available resource contains the full text of more than 30 (predominantly)
molecular biology textbooks. The Bookshelf is available through the main web page
for PubMed: www.PubMed.gov. There is also an entry in the PubMed Bookshelf known
as Coffee Break, a collection of short reports on recent discoveries that cross broad
areas of interest. Most reports demonstrate how new advances in molecular biomedicine
can be analyzed with NLM computer tools to better understand pathophysiology and
lead to new therapies.
Many other government-sponsored resources are available on the
WWW for medical researchers. A complete list would not fit in the space available
for this review and, in any case, would be outdated by publication date. A website
of particular note, however, is that of the NIH (www.nih.gov). Here, in addition
to information on obtaining grants and fellowships is the "CRISP" database, which
provides information on NIH-funded research projects. Other resources at the NIH
website include genetic maps, computational tools for biology, and many technical
documents from other parts of the federal government.
 Figure 80-4
Growth in the use of PubMed. As of November 2003, there
were approximately 2 million PubMed searches per day. (Data courtesy of
PubMed.gov.)
Figure 80-4
Growth in the use of PubMed. As of November 2003, there
were approximately 2 million PubMed searches per day. (Data courtesy of
PubMed.gov.)